Software Description:
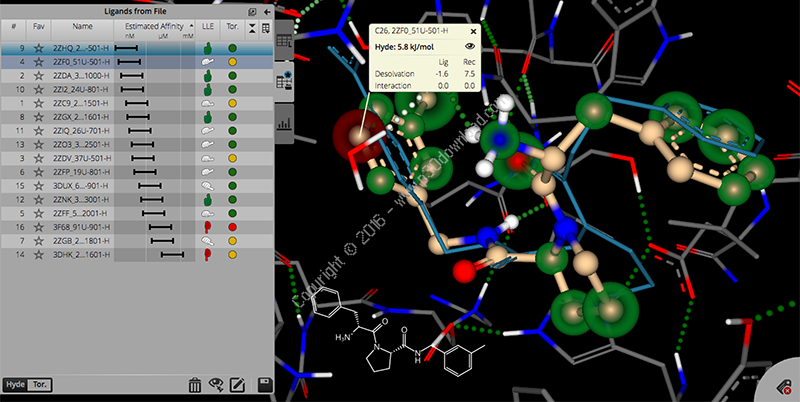
SeeSAR is a software tool for interactive,visual compound prioritization as well as compound evolution.Structure-based design work ideally supports a multi-parameteroptimization to maximize the likelihood of success, rather thanaffinity alone. Having the relevant parameters at hand incombination with real-time visual computer assistance in 3D is oneof the strengths of SeeSAR.
Affinities:
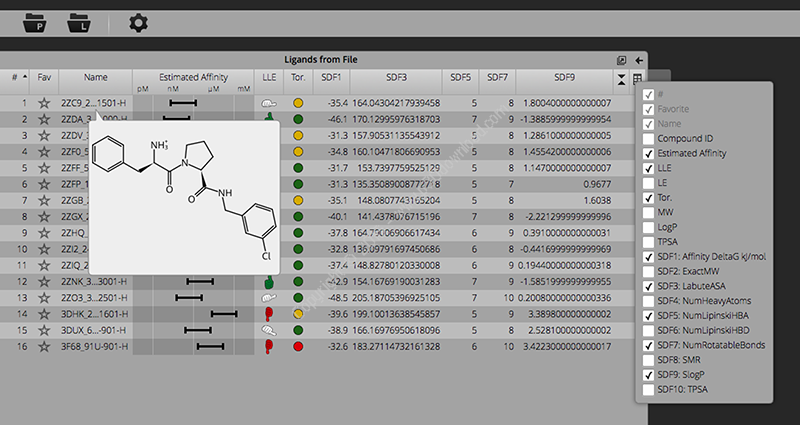
We implemented sophisticated graphics to visualize atom-basedaffinity contributions; that allow for a rough estimate of the ΔS /ΔH -split of the Free Energy. (This is an ongoing co-developmentbetween BAYER, the University of Hamburg and BioSolveIT.)
Phys-chem properties:
Relevant parameters are computed on-the-fly or imported to be takeninto consideration throughout the design process.
Torsional ‘heat’:
Torsional statistics analyses (developed between Hoffmann-LaRocheand the University of Hamburg); is readily available via intuitivecolor-coding.
Explorable space:
A tight fit is the prerequisite for both, affinity and specificity.Therefore, as guidance for the user, efficient computation combinedwith refined graphics provides on-the-fly visualization of gaps inthe binding interface and positions where a tighter fit is likelyto be gained.
2D molecule browsing – time to look at things from adifferent angle!
While the molecule table offers great functionality forprioritizing compounds based on the data, it does not provide anoverview of the molecules themselves. This release, however, seesthe introduction of 2D molecule browsing. The table now offers twoviews – the one you already know and a 2D browser – flick betweenthem using the switch below the table. Both views are always keptin sync so if you add a filter or sort etc. the 2D browser willshow you the same result in the same order as the table. Also tryexpanding the table area to see how more molecules fit into theview.
Fantastic new 3D graphics features
This release also brings with it some great new 3D graphicsimprovements. As much as we all like visualising the binding sitesurface, it lay often times in the way… The binding site surfacecan now be switched to transparent allowing you to see through itand therefore making the analysis of the binding site and moleculeswithin much more comfortable. Also, the feeling of depth in the 3Dview has been improved to help orientation – a so-called “fogeffect” fades out the protein and molecules that are further awayto bring the foreground more into focus.
Persistent amino acid labels and better view ofreference
So far, labels on binding site components unfortunately disappearedwhen browsing through different molecules in the table. Now aminoacid, co-factor and water labels remain present if you change to adifferent molecule in the 3D view and even if you enter themolecule editor. The view of the reference compound has also beenimproved. For better visibility, the thickness of the bonds hasbeen increased and instead of coloring the whole molecule in auniform blue color, only the carbon atoms are colored blue so thathetero atoms can be distinguished more easily.
Installer Size: 29.3 MB
Download Links : SeeSAR v6.1 + Crack